A sneak peak into the recent Antimicrobial Resistance Research and Surveillance Network (AMRSN) 2022 Report
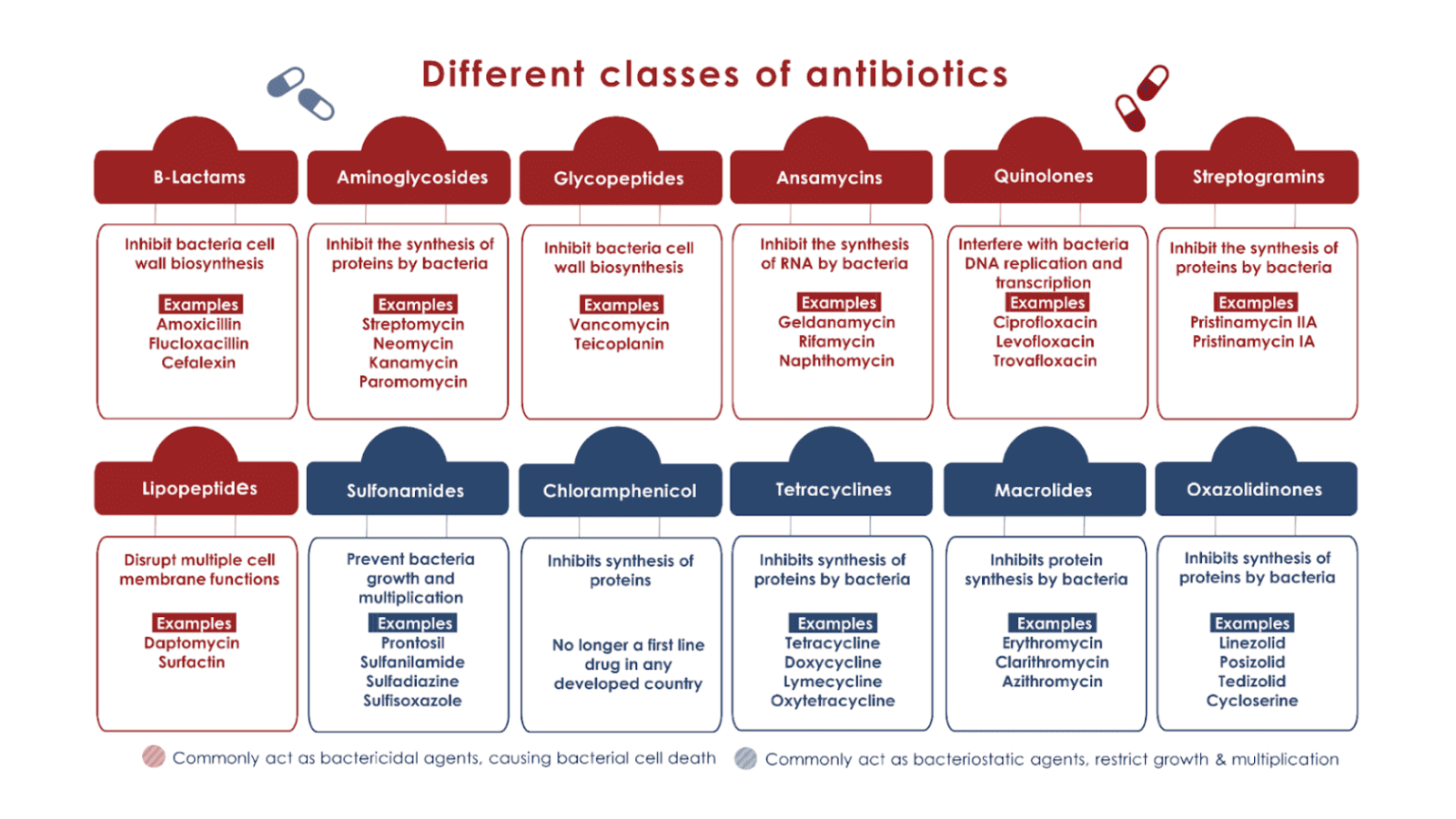
Antimicrobial Resistance (AMR) poses a significant threat to global health, and India is no exception. AMR occurs when microorganisms, such as bacteria, viruses, and fungi, develop resistance to drugs that were initially effective in treating infections caused by them. Since the discovery of penicillin, various antibiotics (see figure below) have been developed over the last several decades to treat specific infections – from mild to severe – and have since transformed modern medicine.
However, lately, the overuse and misuse of antibiotics in healthcare, farming and agriculture have become major contributors to the emergence and spread of AMR which, according to a report released in The Lancet, killed at least 1.27 million people worldwide and is associated with nearly 5 million deaths in 2019 alone.
Antimicrobials, such as antibiotics, are being used excessively, which is accelerating an otherwise normal process of genetic evolution in microorganisms, in which bacteria and other microorganisms develop mutations over time to resist the toxic effects of antimicrobials in order to survive. While genetic mutations driving evolution are a widespread occurrence among diverse species, the fast replication time of microorganisms such as bacteria and their ability to transfer genes within bacterial populations means the resistant genes spread faster. These biological characteristics, combined with poor hygiene and sanitation conditions, as well as rapid globalisation and climate change, ensure that AMR infections spread widely.
Source: ReAct
The same Lancet study also highlighted six disease-causing bacteria types – Escherichia coli (E. coli), Streptococcus pneumoniae (S. pneumoniae.), Klebsiella pneumoniae (K. pneumoniae), Staphylococcus aureus (S. aureus), Acineobacter baumanii (A. baumanii) and Pseudomonas aeruginosa (P. aeruginosa) alone – that alone caused nearly 1 million deaths. Among these, E. coli was the most deadly pathogen, claiming 0.157 million lives in India in 2019. The study also showed that common bacterial infections were the second-leading cause of death in 2019, and were linked to one in eight deaths globally, making it critical to monitor the rise and spread of these ‘superbugs’.
The latest Annual Report by the Antimicrobial Resistance Research and Surveillance Network (AMRSN) reveals critical insights into the state of AMR in India in 2022. The AMRSN, under the aegis of the Indian Council of Medical Research (ICMR), has been working since 2013 to collect, analyse, and understand data related to AMR. This sixth annual report presents a comprehensive overview of AMR trends, patterns, and challenges faced by India’s tertiary healthcare system. However, the report lacks information on the pattern of AMR in Mycobacterium tuberculosis, a pathogen that causes tuberculosis and is resistant to a variety of antibiotics, making its treatment extremely challenging.
The report also highlights the often-overlooked role of genomics and Whole Genome Sequencing (WGS) in understanding resistance mechanisms among key priority pathogens.
Genomic sleuths for tracking infections
As demonstrated during COVID-19, genomics surveillance has evolved into an important tool for monitoring and investigating pathogens and its spread. Recognising the importance of this approach in both local and global surveillance, WHO has developed a 10-year Global genomic surveillance strategy for pathogens with pandemic and epidemic potential 2022 – 2032.
To undertake genomic surveillance, samples are collected from patients, animals, or the environment that may contain the pathogen of interest. A swab from a patient’s throat or nose, a blood sample, or even a sample from contaminated food or water could be used. Following that, DNA/RNA from these samples is extracted and analysed to determine the type of pathogen or, for that matter, whether it is present at all. By doing so, we can learn more about what causes infectious diseases, where pathogens originate, and how they spread. It’s like looking for clues to catch the bad guys (pathogens) and prevent them from causing havoc (outbreaks/pandemic). As a result, genomic surveillance can be a useful tool only if adequate infrastructure is in place to continuously monitor and report comprehensively.
The AMRSN study uses genomic surveillance as well as the analysis of antibiograms* from outpatient departments (OPD), wards, and intensive care units (ICU), which play a critical role in evaluating the trends of AMR and its significance in clinical practice, particularly for the appropriate use of antibiotics, to shed light on the ongoing battle against AMR in India.
*An antibiogram is a report summarising bacteria’s susceptibility or resistance to antibiotics, helping doctors choose effective treatments and monitor antibiotic resistance trends.
Molecular clues from AMRSN Report 2022
The study examined 1,07,053 isolates from blood, urine, faeces, superficial infections, deep infections, lower respiratory tract, sterile sites*, cerebrospinal fluid, and others that tested positive for . E. coli was the most commonly isolated pathogen followed by the K. pneumoniae, P. aeruginosa, A. baumannii, and S. aureus. These organisms are known for their pathogenicity and propensity for developing resistance against antibiotics used against them.
The study looked at ‘antibiotic sensitivity or susceptibility’ which shows if bacteria are susceptible (can be treated with the antimicrobial drug), intermediate (may be treatable with the drug, but may require adjusted dosage), or resistant (cannot be treated with drug).
The report emphasises that the data collected primarily comes from tertiary care hospitals in 21 different locations and does not reflect community-level AMR. Thus, the findings should not be extrapolated to community settings or to the environment.
* ‘Sterile sites’ are areas in the body where microorganisms are typically not found and are often placed deeper in the body and more protected from outside infection. For example, abdominal fluid, pancreatic drain fluid.
The ongoing rise of the superbugs
E. coli and K. pneumoniae are common pathogens isolated from urine from OPD, wards and ICU. The data in the AMRN report shows that susceptibility of E. coli to Imipenem, a commonly used carbapenem antibiotic, decreased from 81% in 2017 to 66% in 2022. Imipenem is used to treat severe bacterial infections. K. pneumoniae showed a similar trend, with Imipenem sensitivity dropping from 59% in 2017 to 42% in 2022.
Carbapenem resistance in A. baumannii, pathogen that commonly responsible for the Urinary Tract Infections (UTIs), penumonia, among others, was alarmingly high at 87.8% in 2022 with minocycline exhibiting the highest susceptibility after colistin, reaching around 58%.
The study found that the ICUs faced high prevalence of AMR, with significant percentages of imipenem resistance in K. pneumoniae and A. baumannii. S. aureus and Enterococcus faecium (E. faecium) causing bloodstream infections (BSIs) in ICUs exhibited resistance to oxacillin and vancomycin, respectively.
Pseudomonas aeruginosa (P. aeruginosa), a common bacterial pathogen that causes a variety of infections, particularly in patients in hospital, showed higher sensitivity to antipseudomonal cephalosporins in ward isolates compared to ICU.
Colistin, known as the last-resort antibiotic, remained highly effective against P. aeruginosa, with no notable change in sensitivity over five years. Colistin is also used extensively in animal and plant farming which is fueling the AMR crisis and resulting in its ineffectiveness in human health.
The AMRN report also noted that the Methicillin-Resistant Staphylococcus aureus (MRSA) rates have been on the rise, increasing from 28.4% in 2016 to 42.6% in 2021. However, antibiotics vancomycin and teicoplanin remained highly effective against MRSA isolates, with Levonadifloxacin showing promise as a potential treatment option for certain infections.
Salmonella Typhi (S. Typhi), the typhoid fever-causing bacterium, exhibited good sensitivity to antibiotics like ceftriaxone, cefixime, trimethoprim-sulfamethoxazole, and azithromycin. However, fluoroquinolones, the primary choice for treating typhoid, showed limited effectiveness against S. Typhi.
In addition to these findings, the AMRSN’s 2022 Annual Report includes data on various other pathogens studied not included in this write-up.
Towards better surveillance
The AMRSN’s 2022 Annual Report reveals that while the patterns of AMR have remained somewhat stable in the last 5-6 years, AMR is still on the rise and this data should provide enough impetus to combat AMR in our country. High rates of resistance in critical care units as reported in this study underscore the urgent need for better infection control measures in hospital settings and a more robust environmental surveillance.
Effective antibiotic stewardship, lab diagnostics, novel antimicrobials, and increased awareness about responsible antibiotic use are essential to combat AMR in India. Continuous genomic surveillance remains an important tool in monitoring and addressing this pressing public health concern. Ultimately, the battle against AMR requires a concerted effort from healthcare professionals, policymakers, and the public to preserve the effectiveness of antibiotics for future generations.
SaS is currently part of Alliance for Pathogen Surveillance India (APSI-India), which is a multi-city consortium to develop advanced pathogen surveillance platforms for detection of disease-causing agents in environmental samples such as wastewater, and monitoring their correlation with clinical caseloads. Find out more about this initiative here.
You can also read on our blog an informative interview with Dr Uma Bala Pamidimukkala on monitoring AMR in healthcare settings.
